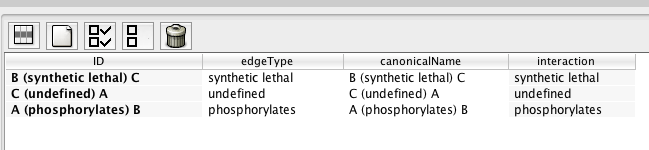
Both Cytoscape and RCytoscape attach special significance to one particular edge attribute: this attribute, in both cases, determines the type of the edge. It is easy to get confused, however, because a different attribute is used in the two enviroments:
interaction
edgeType
edgeType in your R graph.
g <- initEdgeAttribute (graph=g, attribute.name='edgeType', attribute.type='char', default.value='undefined')
For example, if you have code like this in R (from RCytoscape::makeSimpleGraph ())
g = new("graphNEL", edgemode = "directed")
g = graph::addNode("A", g)
g = graph::addNode("B", g)
g = graph::addNode("C", g)
g = initEdgeAttribute(g, "edgeType", "char", "undefined")
g = graph::addEdge("A", "B", g)
g = graph::addEdge("B", "C", g)
g = graph::addEdge("C", "A", g)
edgeData(g, "A", "B", "edgeType") = "phosphorylates"
edgeData(g, "B", "C", "edgeType") = "synthetic lethal"
